| Name | Strains | Location (C57BL/6J) | Clones (C57BL/6J) |
|---|---|---|---|
| Idd5.1 |
NOD/MrkTac, NOD/ShiLtJ |
1:65533102-69307244 | AL683804.15 to AL645534.20 |
| Idd5.3 | NOD/MrkTac | 1:65533102-69307244 | AC100180.12 to AC101699.9 |
| Idd5.4 | NOD/MrkTac | 1:130232728-130661594 | AC123760.9 to AC109283.8 |
| Idd3 |
NOD/MrkTac, 129 |
3:36492618-37600833 | AC117584.11 to AL627074.11 |
| Idd10 | NOD/MrkTac | 3:99848826-101467080 | AC167172.3 to AC131184.4 |
| Idd18.1 | NOD/MrkTac | 3:109144756-109930492 | AL845310.4 to AL683824.8 |
| Idd18.2 | NOD/MrkTac | 3:103489414-104054885 | AC123057.4 to AC129293.9 |
| Idd9.1 | NOD/MrkTac | 4:128371876-131853368 | AL627093.17 to AL670959.8 |
| Idd9.1M | NOD/MrkTac | 4:134841437-135252443 | AL611963.24 to AL669936.12 |
| Idd9.2 | NOD/MrkTac | 4:146049976-149895141 | CR788296.8 to AL626808.28 |
| Idd9.3 | NOD/MrkTac | 4:149556939-151385803 | AL607078.26 to AL606967.14 |
| Idd6.1 + Idd6.2 (combined) | NOD/ShiLtJ | 6:143550839-149565172 | AC164704.4 to AC164090.3 |
| Idd6.AM | NOD/ShiLtJ | 6:129593784-131241919 | AC171002.2 to AC163356.2 |
| Idd4.1 | NOD/MrkTac | 11:69704895-71153537 | AL596185.12 to AL663042.5 |
| Idd4.2 | NOD/MrkTac | 11:72734492-74404570 | AL663082.5 to AL604065.7 |
| Idd4.2Q | NOD/ShiLtJ | 11:86785996-90007691 | AL596111.7 to AL645695.18 |
| Idd16.1 | NOD/ShiLtJ | 17:27302611-29220265 | AC125141.4 to AC167363.3 |
| Idd1 (MHC) |
NOD/MrkTac, NOD/ShiLtJ |
17:33567721-38721962 | CT033847.5 to AC134471.3 |
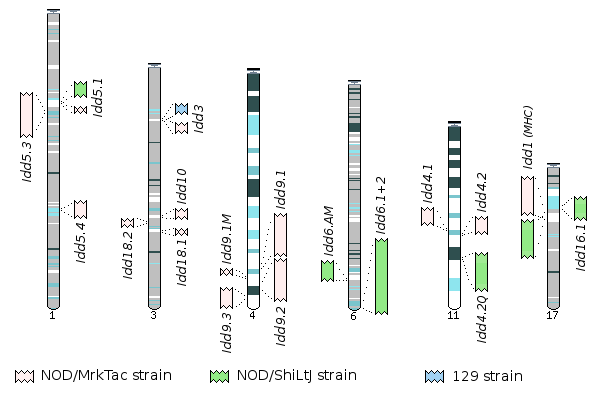
Insulin Dependent Diabetes (Idd) regions in different strains
Vega presents the annotation of several candidate Insulin Dependent (Type 1) Diabetes (Idd) susceptibility loci mapped to chromosomes 1, 3, 4, 6, 11 and 17. These have been annotated as part of a project to sequence the Idd regions in the Non-Obese Diabetic (NOD) mouse genome and to investigate the role of these regions in Type 1 diabetes. They have have been annotated in the reference (C57BL/6J) and one or more of NOD/MrkTac, NOD/ShiLtJ, and 129 strains. This approach allows for the comparison of the genomic sequence and genes in the candidate loci between diabetes resistant and diabetes sensitive strains, looking for example for SNPs (Wicker et al. (2004) J Immunology 173: 164-173). For more information on this project please email Charles Steward or see the NOD mouse website.
The mouse MHC region in C57BL/6J is defined by markers D17Mit16 and D17Mit176 and contains the genes associated with the classic MHC. The homologous region of the MHC is also represented in both NOD mouse strains (NOD/MrkTac and NOD/ShiLtJ) although the NOD/ShiLtJ sequence has been extended to include the olfactory receptors at the distal end.
The regions and strains used are indicated on the table to the right. The figure below shows the approximate locations of these regions on the reference strain and the relative sizes with respect to each other. Clicking on a region in the figure or a link in the rightheand column of the table will take you to the appropriate chromosome summary page.
Further information on NOD mouse genome annotation can be found in:
-
The non-obese diabetic mouse sequence, annotation and variation resource: an aid for investigating type 1 diabetes.
Steward CA, Gonzalez JM, Trevanion S, Sheppard D, Kerry G, Gilbert JG, Wicker LS, Rogers J and Harrow JL
Database : the journal of biological databases and curation 2013;2013;bat032 PUBMED: 23729657; PMC: 3668384; DOI: 10.1093/database/bat032

